
You will need this user to set up a secure connection to the restored Creatio database using PostgreSQL authentication.
This user will restore the Creatio database from a backup file and assign access permissions.Ī user with the “ public” role, whose permissions are limited. We recommend to create two database users in PostgreSQL:Ī user with the “ sysadmin” role, who has maximum access privileges on the database server level. By default, no such user will be available.
#Postgresql database update#
If you plan to use a fresh installation of PostgreSQL, you need to create a user that can log in to the database using a password and has sufficient privileges to create and update a database. Create PostgreSQL userĪ fresh installation of PostgreSQL is not ready for deploying Creatio immediately.
#Postgresql database code#
The public Postgres database allows anyone to run the RNAcentral website locallyĪnd contribute new code to RNAcentral using the instructions available on GitHub.Note. Please refer to the PostgreSQL documenation for details on PostgreSQL clustering.
#Postgresql database install#
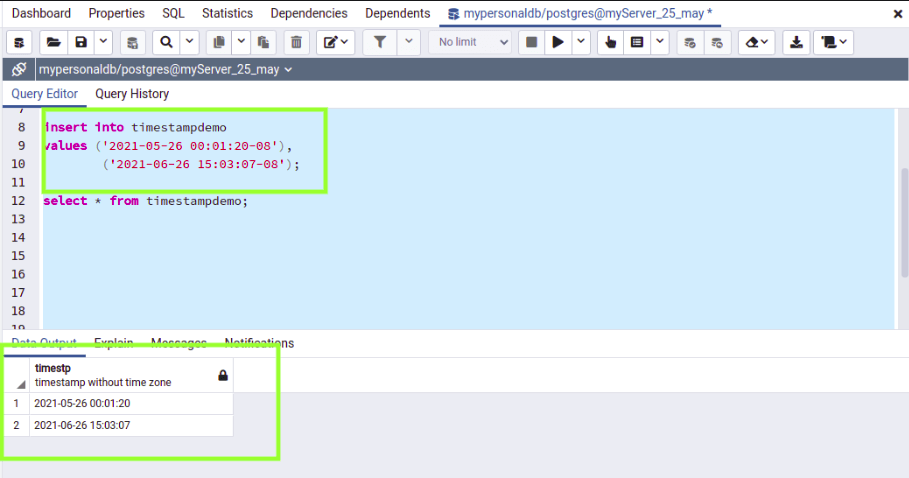
Requires psycopg2 to connect to Postgres: pip install psycopg2 The file output.fasta will contain the desired subset of RNAcentral sequences in FASTA format. The command will create a file ids.txt with a list of RNAcentral identifiers.ĭownload the following RNAcentral FASTA file:Įxtract the sequences using seqkit: seqkit grep -f ids.txt rnacentral_species_specific_ > output.fasta Run the following command to execute the query: docker run -v `pwd`:/rnacentral -it postgres /bin/sh -c 'cd /rnacentral & psql -t -A -f query.sql > ids.txt' Tax.lineage LIKE 'cellular organisms Bacteria %'ĪND precomputed.is_active = true - exclude sequences without active cross-references If you need to export more sequences, you can use the following workflow:

The RNAcentral text search can only export up to 1,000,000 search results. You can still query RNAcentral using VEGA identifiers: SELECT Identifiers are no longer searchable using the RNAcentral text search, rnc_rna_precomputed - contains RNA types and descriptions for all sequencesĮxample queries Search by external accessionsĪlthough the VEGA database has been archived and its.rnc_accessions - contains metadata associated with each cross-reference.rnc_database - contains a list of Expert Databases.xref - contains cross-references to Expert Databases.rna - contains RNA sequences and URS identifiers.The entire RNAcentral schema contains more than 40 tables, but the following tablesĪre good starting points for exploring the data: The following diagram was generated based on RNAcentral release 20. If your computer is behind a firewall, please ensure that outgoing TCP/IP connections to the corresponding ports are allowed. Psql tip: if you don't have psql installed on your machine, consider using Docker to get started with a pre-configured Postgres image:ĭocker run -it postgres psql you can use a Postgres client like DBeaver or PgAdmin. To connect to the database using command line: The database is updated with every RNAcentral releaseĪnd contains a copy of the data available through the RNAcentral website.
In addition to downloadable files, an API,Īnd the text search, RNAcentral provides a public


 0 kommentar(er)
0 kommentar(er)
